Projects
AI4Wildlive

The WildLIVE portal is a data management platform for wildlife monitoring data. The goal is to create an integrated platform for managing and analyzing camera trap data. The platform incorporates AI-based image annotation to assist in identifying species captured in camera trap images, human curation to validate and improve annotations, FAIR data principles to ensure data accessibility and reusability.
More information about AI4Wildlive can be found on the FEDA project page.
A FAIR workflow platform

Deployed platform: https://workflow.earth
Poster DOI: 10.5281/zenodo.13928655
FAIR stands for Findable, Accessible, Interoperable, Reuseable. By making research data available in a FAIR manner, researchers can make their data more useful! However, implementing FAIR principles is still challenging.
This platform provides a practical implementation of FAIR principles for computational workflows. It executes machine-actionable workflows while capturing complete provenance information, packaging both workflows and results as RO-Crates. This ensures that scientific workflows and their outputs remain findable, accessible, interoperable, and reusable throughout their lifecycle.
The platform is currently deployed within the Destination Earth Data Lake where it drives BioDT’s Digital Twin Model for finding crop wild relatives of plants for enhancing crop resilience against climate-driven stresses. This use case helps identify stress-tolerant populations and novel genetic traits for crop improvement (more info).
The platform code is open-source and available on Github. A poster describing the project details is available at the DOI linked above.
griesheim-transparent.de
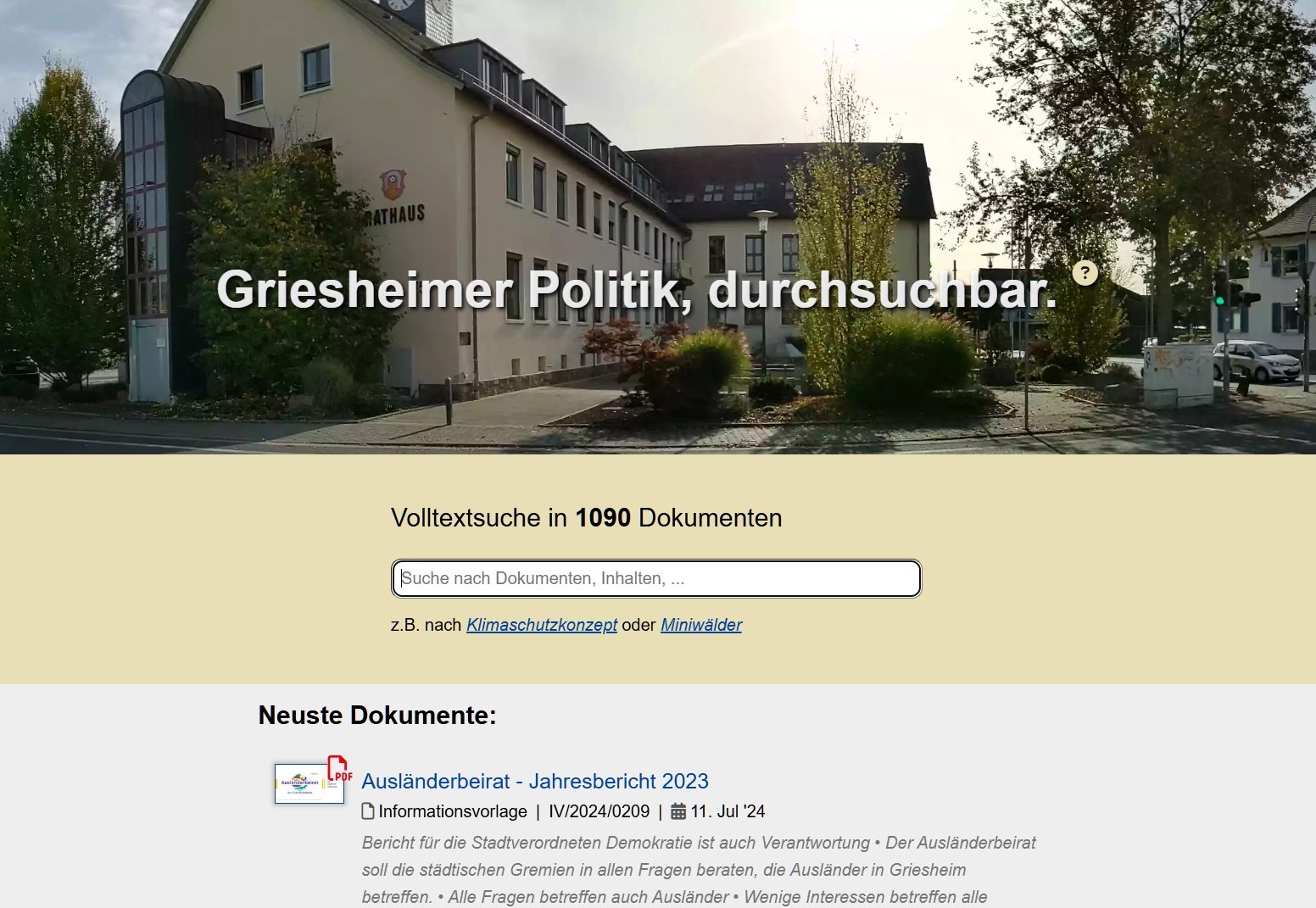
https://griesheim-transparent.de is a platform designed to increasing transparency in local government for the citizens of Griesheim.
The platform indexes documents from the local government and makes them available through a search interface. It provides full-text search capabilities on resolutions, council meeting minutes, etc.
The goal is to empowers citizens by enabling easy access to important governmental documents and fostering greater community involvement.
Corax: A Split ergo keyboard

The Corax is a custom mechanical keyboard I designed and built from the ground up. It is fully programmable using the ZMK firmare, featuers a split and column-staggered layout, bluetooth, and hot-swappable switches.
The design of the PCB, 3D printed parts and the firmware are available on Github.
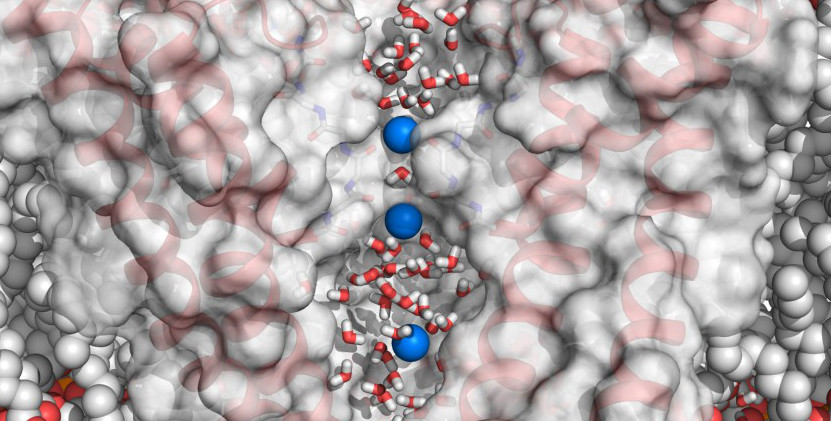
Molecular dynamics simulations of ion channels
DOI: 10.26083/tuprints-00018611
During my PhD, I focused on understanding the workings of ion channels, particularly HCN channels. These channels are essential in regulating heart rhythms and various neuronal processes in human.
I used molecular dynamics simulations to explore ion conductance properties and contributed to research related to channelopathies (diseases related to channel malfunctioning).
Weighted Histogram Analysis Method (WHAM)
wham --max 3.14,3.14 --min -3.14,-3.14 -T 300 --bins 100,100 --cyclic -f example/2d/metadata.dat
> Supplied WHAM options: Metadata=example/2d/metadata.dat, hist_min=[-3.14, -3.14], hist_max=[3.14, 3.14], bins=[100, 100] verbose=false, tolerance=0.000001, iterations=100000, temperature=300, cyclic=true
> Reading input files.
> 625 windows, 624262 datapoints
> Iteration 10: dF=0.389367172324539
> Iteration 20: dF=0.21450559607810152
(...)
> Iteration 620: dF=0.0000005800554892309461
> Iteration 630: dF=0.00000047424278621817084
> Finished. Dumping final PMF
(... pmf dump ...)
The Weighted Histogram Analysis Method (WHAM) is a fast implementation of WHAM written in Rust. It allows the calculation of multidimensional free energy profiles from umbrella sampling simulations. I wrote WHAM during my PhD because there was no polished implementation of WHAM that was able to utilize more than one CPU. This implementation is particularly well-suited for larger systems and and features additional error analysis features.